1. Introduction¶
Metacat is a repository for data and metadata (descriptions of data) that helps scientists find, understand and effectively use the data sets they manage or that have been created by others. Thousands of data sets are currently documented in a standardized way and stored in Metacat systems, providing the scientific community with a broad range of science data that–because the data are well and consistently described–can be easily searched, compared, merged, or used in other ways.
Not only is the Metacat repository a reliable place to store metadata and data (the database is replicated over a secure connection so that every record is stored on multiple machines and no data is ever lost to technical failures), it provides a user-friendly interface for information entry and retrieval. Scientists can search the repository via the Web using a customizable search form. Searches return results based on user-specified criteria, such as desired geographic coverage, taxonomic coverage, and/or keywords that appear in places such as the data set’s title or owner’s name. Users need only click a linked search result to open the corresponding data-set documentation in a browser window and discover whom to contact to obtain the data themselves (or how to immediately download the data via the Web).
Metacat’s user-friendly Registry application allows data providers to enter data set documentation into Metacat using a Web form. When the form is submitted, Metacat compiles the provided documentation into the required format and saves it. Information providers need never work directly with the XML format in which the metadata are stored or with the database records themselves. In addition, the Metacat application can easily be extended to provide a customized data-entry interface that suits the particular requirements of each project. Metacat users can also choose to enter metadata using the Morpho application, which provides data entry wizards that guide information providers through the process of documenting each data set.
The metadata stored in Metacat includes all of the information needed to understand what the described data are and how to use them: a descriptive data set title; an abstract; the temporal, spatial, and taxonomic coverage of the data; the data collection methods; distribution information; and contact information. Each information provider decides who has access to this information (the public, or just specified users), and whether or not to upload the data set itself with the data documentation. Information providers can also edit the metadata or delete it from the repository, again using Metacat’s straightforward Web interface.
Metacat is a Java servlet application that runs on Linux and Mac OS platforms in conjunction with a database, such as PostgreSQL, a Web server, and a Solr server. The Metacat application stores data in an XML format using Ecological Metadata Language (EML) or other metadata standards such as ISO 19139 or the FGDC Biological Data Profile. For more information about Metacat or for examples of projects currently using Metacat, please see http://knb.ecoinformatics.org.
1.1. What’s in this Guide¶
This Administrator’s guide includes information for installing, configuring, managing and extending Metacat for Ubuntu and other Linux variants. Metacat can run on MacOS but it needs users’ customized installation. Chapter Four contains instructions for downloading and installing Metacat and the applications required to run the software on Linux platform. Chapter Five covers how to configure Metacat, both for new and upgraded installations. Chapter Seven details the ways in which you can customize the Metacat interface so users can access and submit information easily: using Metacat’s generic web-interface (the Registry), creating your own HTML forms, and creating your own desktop client (like Morpho). Chapter Eight discusses how to work with Metacat’s embedded Geoserver. Chapter Nine describes how to set up the Metacat’s replication service, which permits Metacat servers to share data with each other, effectively backing up metadata and data files. Chapter Ten looks at the Metacat Harvester, a program that automates the retrieval of EML documents from one or more sites and their subsequent upload (insert or update) to Metacat. Chapter Eight discusses logging, Chapter Twelve contains instructions for creating a site map, which makes individual metadata entries available via Web searches. Metacat’s Java API is available for developers.
1.2. Metacat Features¶
Metacat is a repository for data and metadata (documentation about data), that helps scientists find, understand and effectively use the data sets they manage or that have been created by others. Specifically,
- Metacat is an open source web application, which can run on Linux and MacOS operating systems and is written in Java
- Metacat’s Web interface facilitates the input and retrieval of data
- Metacat’s optional mapping functionality enables you to query and visualize the geographic coverage of stored data sets
- Metacat’s replication feature ensures that all Metacat data and metadata is stored safely on multiple Metacat servers
- The Metacat interface can be easily extended and customized via Web forms, themes, and/or user-developed client tools in Java and other languages
- The Metacat harvester automates the process of retrieving and storing EML documents from one or more sites
- Metacat can be customized to use Life Sciences Identifiers (LSIDs), uniquely identifying every data record
- Metacat has a built-in logging system for tracking events such as document insertions, updates, deletes, and reads
- The appearance of Metacat’s Web interface can be customized via themes.
- Metacat fully supports the DataONE Member Node interface, allowing Metacat deployments to easily participate in the DataONE federation
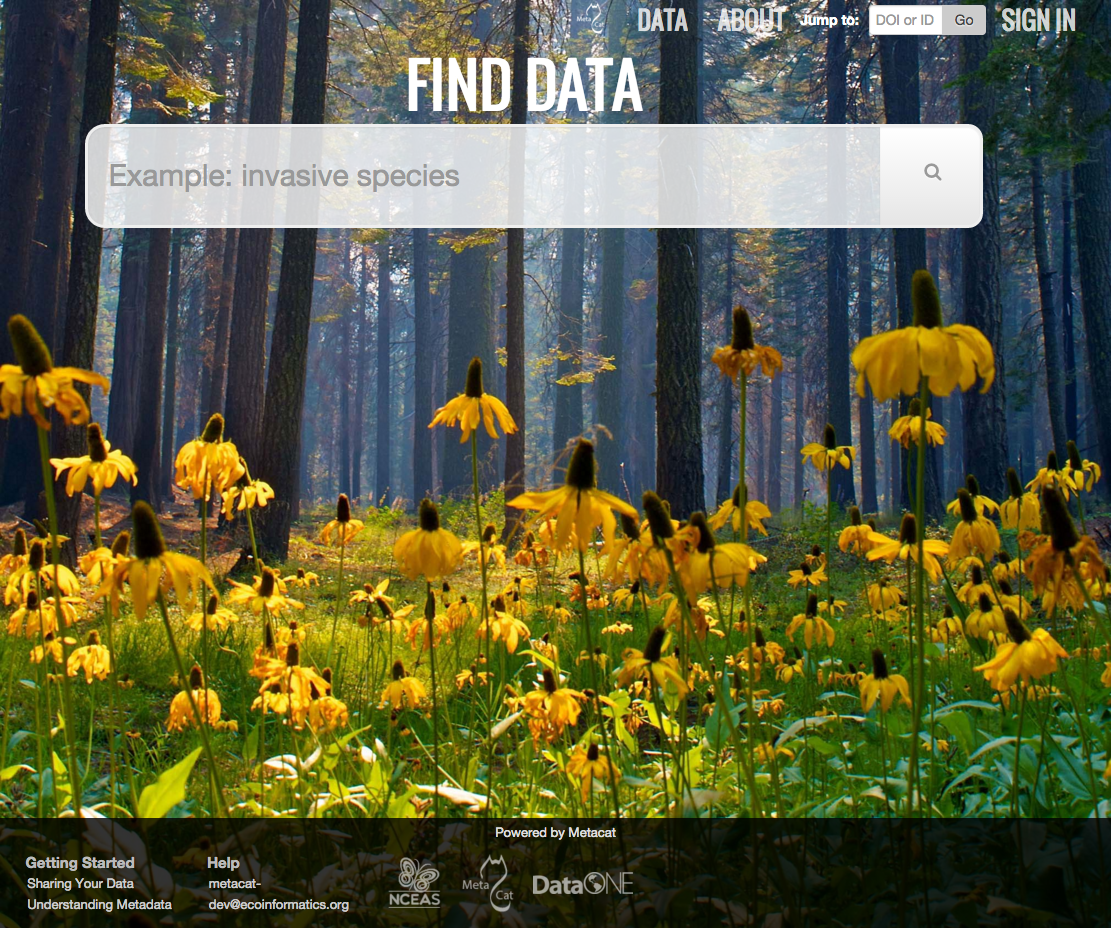
Metacat’s default home page. Users can customize the appearance using themes.